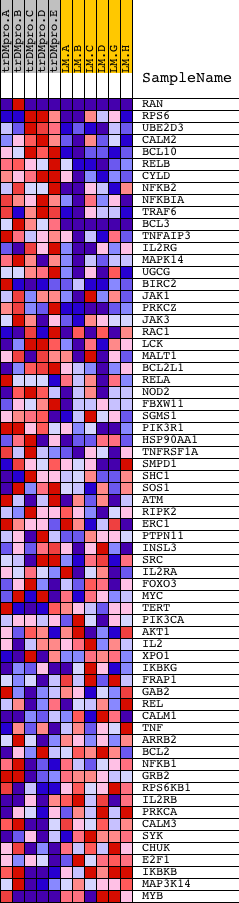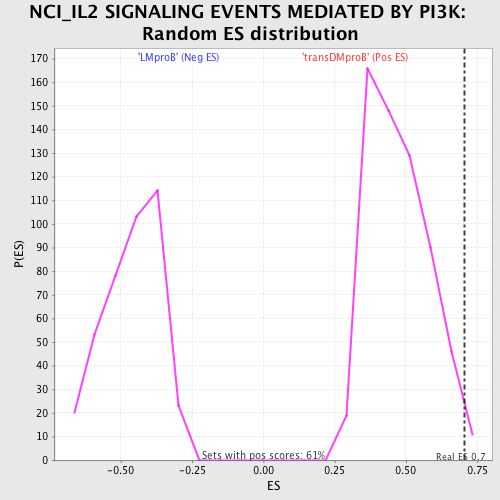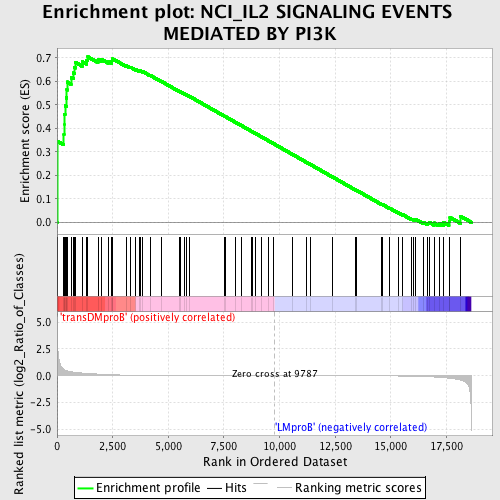
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | NCI_IL2 SIGNALING EVENTS MEDIATED BY PI3K |
| Enrichment Score (ES) | 0.70402235 |
| Normalized Enrichment Score (NES) | 1.4889116 |
| Nominal p-value | 0.01642036 |
| FDR q-value | 0.57008153 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RAN | 5356 9691 | 6 | 4.449 | 0.3457 | Yes | ||
| 2 | RPS6 | 9757 | 306 | 0.562 | 0.3732 | Yes | ||
| 3 | UBE2D3 | 7253 | 312 | 0.557 | 0.4163 | Yes | ||
| 4 | CALM2 | 8681 | 316 | 0.552 | 0.4591 | Yes | ||
| 5 | BCL10 | 15397 | 358 | 0.510 | 0.4965 | Yes | ||
| 6 | RELB | 17942 | 412 | 0.466 | 0.5299 | Yes | ||
| 7 | CYLD | 18532 | 439 | 0.449 | 0.5634 | Yes | ||
| 8 | NFKB2 | 23810 | 451 | 0.442 | 0.5972 | Yes | ||
| 9 | NFKBIA | 21065 | 629 | 0.362 | 0.6158 | Yes | ||
| 10 | TRAF6 | 5797 14940 | 736 | 0.330 | 0.6357 | Yes | ||
| 11 | BCL3 | 8654 | 778 | 0.320 | 0.6584 | Yes | ||
| 12 | TNFAIP3 | 19810 | 814 | 0.314 | 0.6809 | Yes | ||
| 13 | IL2RG | 24096 | 1125 | 0.254 | 0.6839 | Yes | ||
| 14 | MAPK14 | 23313 | 1325 | 0.221 | 0.6904 | Yes | ||
| 15 | UGCG | 16197 | 1383 | 0.214 | 0.7040 | Yes | ||
| 16 | BIRC2 | 4397 4398 | 1839 | 0.152 | 0.6913 | No | ||
| 17 | JAK1 | 15827 | 1982 | 0.134 | 0.6941 | No | ||
| 18 | PRKCZ | 5260 | 2295 | 0.104 | 0.6854 | No | ||
| 19 | JAK3 | 9198 4936 | 2453 | 0.091 | 0.6840 | No | ||
| 20 | RAC1 | 16302 | 2467 | 0.090 | 0.6904 | No | ||
| 21 | LCK | 15746 | 2503 | 0.088 | 0.6953 | No | ||
| 22 | MALT1 | 6274 | 3123 | 0.053 | 0.6660 | No | ||
| 23 | BCL2L1 | 4440 2930 8652 | 3290 | 0.046 | 0.6607 | No | ||
| 24 | RELA | 23783 | 3534 | 0.038 | 0.6506 | No | ||
| 25 | NOD2 | 6384 | 3697 | 0.034 | 0.6445 | No | ||
| 26 | FBXW11 | 20926 | 3738 | 0.033 | 0.6449 | No | ||
| 27 | SGMS1 | 23698 | 3849 | 0.031 | 0.6414 | No | ||
| 28 | PIK3R1 | 3170 | 4187 | 0.024 | 0.6251 | No | ||
| 29 | HSP90AA1 | 4883 4882 9131 | 4705 | 0.018 | 0.5986 | No | ||
| 30 | TNFRSF1A | 1181 10206 | 5485 | 0.012 | 0.5575 | No | ||
| 31 | SMPD1 | 18145 2658 | 5560 | 0.012 | 0.5544 | No | ||
| 32 | SHC1 | 9813 9812 5430 | 5728 | 0.011 | 0.5463 | No | ||
| 33 | SOS1 | 5476 | 5800 | 0.010 | 0.5433 | No | ||
| 34 | ATM | 2976 19115 | 5945 | 0.010 | 0.5363 | No | ||
| 35 | RIPK2 | 2528 15935 | 7512 | 0.005 | 0.4522 | No | ||
| 36 | ERC1 | 1013 17021 995 1136 | 7588 | 0.004 | 0.4485 | No | ||
| 37 | PTPN11 | 5326 16391 9660 | 8014 | 0.003 | 0.4259 | No | ||
| 38 | INSL3 | 9181 | 8268 | 0.003 | 0.4124 | No | ||
| 39 | SRC | 5507 | 8725 | 0.002 | 0.3880 | No | ||
| 40 | IL2RA | 4918 | 8763 | 0.002 | 0.3862 | No | ||
| 41 | FOXO3 | 19782 3402 | 8916 | 0.002 | 0.3781 | No | ||
| 42 | MYC | 22465 9435 | 9167 | 0.001 | 0.3647 | No | ||
| 43 | TERT | 21604 | 9489 | 0.001 | 0.3475 | No | ||
| 44 | PIK3CA | 9562 | 9725 | 0.000 | 0.3348 | No | ||
| 45 | AKT1 | 8568 | 10560 | -0.001 | 0.2900 | No | ||
| 46 | IL2 | 15354 | 11212 | -0.003 | 0.2551 | No | ||
| 47 | XPO1 | 4172 | 11397 | -0.003 | 0.2454 | No | ||
| 48 | IKBKG | 2570 2562 4908 | 12392 | -0.005 | 0.1922 | No | ||
| 49 | FRAP1 | 2468 15991 | 13428 | -0.009 | 0.1371 | No | ||
| 50 | GAB2 | 1821 18184 2025 | 13442 | -0.009 | 0.1371 | No | ||
| 51 | REL | 9716 | 14596 | -0.015 | 0.0761 | No | ||
| 52 | CALM1 | 21184 | 14639 | -0.015 | 0.0750 | No | ||
| 53 | TNF | 23004 | 14931 | -0.019 | 0.0608 | No | ||
| 54 | ARRB2 | 20806 | 15350 | -0.025 | 0.0402 | No | ||
| 55 | BCL2 | 8651 3928 13864 4435 981 4062 13863 4027 | 15543 | -0.029 | 0.0321 | No | ||
| 56 | NFKB1 | 15160 | 15945 | -0.043 | 0.0139 | No | ||
| 57 | GRB2 | 20149 | 16030 | -0.047 | 0.0130 | No | ||
| 58 | RPS6KB1 | 7815 1207 13040 | 16106 | -0.051 | 0.0129 | No | ||
| 59 | IL2RB | 22219 | 16481 | -0.074 | -0.0015 | No | ||
| 60 | PRKCA | 20174 | 16649 | -0.088 | -0.0036 | No | ||
| 61 | CALM3 | 8682 | 16715 | -0.094 | 0.0002 | No | ||
| 62 | SYK | 21636 | 16943 | -0.118 | -0.0028 | No | ||
| 63 | CHUK | 23665 | 17182 | -0.150 | -0.0039 | No | ||
| 64 | E2F1 | 14384 | 17360 | -0.180 | 0.0005 | No | ||
| 65 | IKBKB | 4907 | 17615 | -0.229 | 0.0046 | No | ||
| 66 | MAP3K14 | 11998 | 17656 | -0.238 | 0.0209 | No | ||
| 67 | MYB | 3344 3355 19806 | 18127 | -0.395 | 0.0264 | No |